BioChem
1 Acid-Base
1.1 Terminology
\[ \overset{acid}{HA} + H_2O \rightleftharpoons H_3O^+ + \overset{base}{A^-}\]
- p_ in front of pKa,pKb,pH,pOH always means -log(..)
- pH + pOH = 14
- pH = -log([H+])
- pOH = -log([OH-])
- pKa + pKb = 14
- pKa = -log(Ka)
- pKb = -log(Kb)
- pH + pOH = 14
- Ka acid dissociation constant aka strength of acid
- pI is the pH when protein has no net electrical charge.
1.2 Problem Set
Triprotic acid, HA, Ka1 is 10-2, Ka2 is 10-6 and Ka3 is 10-10 . THe pH range in which H3A- is the predominant form is a pH between?
- 3 and 5
A unknown weak acid, HA has a Ka of 10-5. If 0.1 mole of this acid is dissolved in one liter of water, the percentage of acid dissoaciated at equilibrium is closest to?
\[\displaylines{ [HA] = 0.1M \\ \overset{0.1-x}{HA} \rightleftharpoons \overset{x}{H} + \overset{x}{A} \\ 1 \times 10^{-5} = \frac{[H][A]}{[HA]} \\ 1 \times 10^{-5} = \frac{x^2}{0.1} \\ x = 10^{-3} }\]
What is the pKa for trimethylammonium in water if the base ionization constant (Kb) for trimethylamine is 7.4*10-5 ?
pKa + pKb = 14
pKb = -log(7.4*10-5) = 4.13
pka = 14 - 4.13 = 9.87
DNA polymerase contains a lysine residue that is important for binding o DNA. Mutations were found that converted this lysine to either glutamate, glycine, valine or arginine. Which mutations would be predicted to be the most and least harmful to the ability of the enzyme to bind DNA?
Arginine is least harmful b/c basic like Lysine and ionizable.
Glycine is most harmful b/c nonpolar meaning it will not bind to DNA.
The quantitative differences in biological activity between the two enantiomers of a compound are sometimes quite large. For example, the D isomer of the drug isoproterenol, used to treat mild asthma, is 50 to 80 times more effective as a bronchodilator than the L isomer. Identify the Chiral center is isoproterenol. Why might 2 enantiomers have such radically different bioactivity?
Because they cannot act on the same binding site
A Peptide has the sequence: Glu-His-Trp-Ser-Gly-Leu-Arg-Pro-Gly
a) Draw the structure of peptide and state it’s single letter sequence
b) What is the net charge of the molecule at pH 3, pH 8, pH 11
c) Estimate the pI for this peptide
| pH | N-Term | Glu(E) | His(H) | Trp(Y) | Ser(S) | Gly(G) | Leu(L) | Arg(R) | Pro(P) | Gly(G) | C-term | Net |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | +1 | 0 | +1 | - | - | - | +1 | - | - | -0.5 | +2.5 | |
| 8 | +0.5 | -1 | 0 | - | - | - | +1 | - | - | -1 | -0.5 | |
| 11 | 0 | -1 | 0 | - | - | - | +1 | - | - | -1 | -1 | |
| (Deduced)6 | +1 | -1 | +0.5 | - | - | - | +1 | - | - | -1 | +0.5 |
Even though we we never asked to write out the row of charges for pH 6,
we noticed that the net charge of 0 must be between pH 3 and pH 8, so we make a guess that pH 6 should be where Net Charge may be close to 0.
But we were off since it’s pH 6 has a Net Charge of +0.5.
Therefore another deduction is \(pI=\frac{6+8}{2}=7\) , meaning pI for this peptide is 7.
Histones are proteins found in eukaryotic cell nuclei, tightly bound to DNA, which has many phosphate groups. The pI of histones is very high, about 10.8. What amino acid residues must be present in relatively large numbers in histones? In what way do these residues contribute to the strong binding site of histones to DNA?
arginine b/c the charge creates ionic interactions
2 Tm and Syn-Anti
2.1 Problem set
Explain the Tm trend in Kool’s P phi experiments. Account for the Tm differences between rows 3,4,8,9,10, 12. The trend is as follows in order of decreasing Tm:
A-T > P-P > P-phi > A-P >> A-phi
A-T has a high Tm b/c it is a base pair that hydrogen bonds.
P-P is second because there is steric hinderence but they stack on each other and have hydrophobic interactions.
P-phi is third because the P is unpaired but more hydrophobic and larger than A.
A-P is fourth b/c it is steric
The Tm of a certain 10mer DNA duplex is typically 50C. In the presence of an intercalating dye, ethidium, the structure of which is shown below, the Tm increases to 70C. Explain
The dye stabilizes the DNA in between the spaces of the base pair through hydrophobic interaction.
Which confirmation of A exists in B-DNA?

Anti because the hydrophilic phosphate group needs to face the outside
while the hydrophobic base pair will be more stable inside, closer to it’s matching base pair.
The drug cordycepin is 3’-dexyadenosine. In vivo the drug is converted to the triphosphate and inhibits RNA synthesis. How might the drug accomplish this? (figure shown)
The drug will bind as a base pair and end the sequence since there is no Hydroxyl group on the 3’ end.
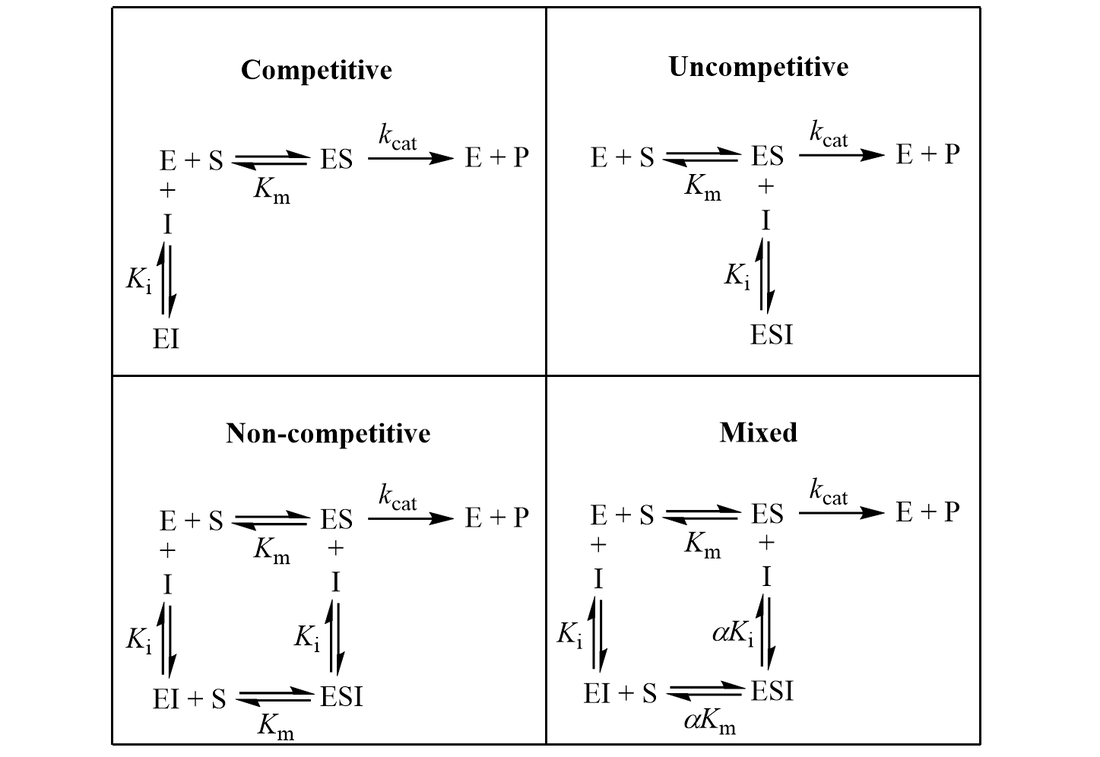
3 Enzymes and Inhibition
General Equation for Inhibition is given by:
\[v = V_{max} \frac{[S]}{K_M \alpha + [S] \beta }\]
| Type | \(\alpha\) | \(\beta\) | Eqn |
|---|---|---|---|
| Mixed Inhibition | \(\alpha = 1 + \frac{I}{K_i}\) | \(\beta = 1 + \frac{[I]}{K_i'}\) | |
| Competitive Inhibition | \(\alpha = 1 + \frac{I}{K_i}\) | \(\beta = 1\) | |
| UnCompetitive Inhibition | \(\alpha = 1\) | \(\beta = 1 + \frac{[I]}{K_i'}\) | |
| NonCompetitive Inhibition | \(\alpha = 1 + \frac{I}{K_i}\) | \(\beta = 1 + \frac{I}{K_i}\) |

4 10
- Mushroom toxin cordycepin is an inhibitor of transcription. Antibiotic rifamycin blocks transcription in bacteria.
5 Proteins
5.1 Concepts
- Proteins are chains of amino acids
- Amino acids have a N-terminus, C-terminus thus Proteins also have this
- N-terminus is a free amine group aka amino terminus
- C-terminus is a free caryboxylic group aka carboxyl terminus
5.2 Terminology
- Monoclonal antibody - Antibodies that bind to one specific epitope
- His-tag - A tag that can be used in affinity chromatography so that other particles can elute first
- Blosum 62 - A system to gauge conservation of amino acid sequences
- Chargaff’s Rules - Rules that states “% moles of A = T” and “% moles of G = C”
5.3 Problem Set
5.3.1 Q1
Propose a quaternary structure for protein complex formed from different subunits with squares representing proteins. Connect them with lines representing disulfide bonds.
+ The mass of the protein complex is 240 kD ( 1 kD=1000 times mass of H) determined by sedimentation velocity.
+ On SDS PAGE, 2 bands are seen, one with MW=200kD, second with MW=10kD.
+ On SDS PAGE, after treated with mercaptoethanol, 3 bands are seen, one at 80kD, one at 30kD, one at 10kD
+ After treating the native protein with increasing concentrations of a bifunctional cross-linking reagent like glutaraldehyde, which covalently joins lysine residues that are close in space, the products show new bands on SDS PAGE, at 210kD, 220 kD, 230 kD, 240kD. The original bands at 200kD and 10kD decrease in intensity.
5.3.2 A1
Solution:
+--+ +--+ +--+ +--+
|10| |10| |10| |10|
+-+--+-+ +-+--+-+ +-+--+-+ +-+--+-+
| 80 +-S--S-| 80 +-S--S-| 30 +-S--S-| 10 |
| | | | | | | |
+------+ +------+ +------+ +------+Mass of the protein complex (240 kD): The total mass of the complex is 240 kD.
SDS PAGE - Untreated (without reducing agent):
Two bands appear at 200 kD and 10 kD. This suggests that before reducing treatment, there is a large complex (likely made of multiple subunits) that gives a band at 200 kD, and a small subunit giving a band at 10 kD. SDS PAGE - Treated with mercaptoethanol (reducing agent):
After reduction of disulfide bonds, three distinct bands appear at 80 kD, 30 kD, and 10 kD. The band at 10 kD persists, suggesting it is a stable small subunit. The new bands at 80 kD and 30 kD imply that the original 200 kD complex is composed of these subunits held together by disulfide bonds. Cross-linking with glutaraldehyde:
The appearance of bands at 210 kD, 220 kD, 230 kD, and 240 kD, with the original 200 kD and 10 kD bands decreasing, suggests that the subunits in the 200 kD complex are closely positioned and are being cross-linked progressively by the reagent. This cross-linking stabilizes different intermediate stages of the assembly.
5.3.3 Q2
Bacillus brevis contains a peptide with antibiotic properties. This peptide disrupts ion transport across the cell membranes of other bacterial species, killing them. Determine the primary structure of the peptide given the following observations.
+ Complete acid hydrolysis of the peptide followed by amino acid analysis yielded equimolar amounts of Leu, Lys, Phe, Pro and Val.
a) Molecular weight of the peptide was estimated to be about 1100 daltons (assume residues have avg molecular weight of 110 daltons)
b) The peptide failed to undergo hydrolysis when treated with the enzyme carboxypeptidase. This enzyme catalyzes the hydolysis of the carboxy-terminal residue of a polypeptide unless the residue is Pro or does not contain a free carboxyl group.
c) Edman degradation did not yield any PTH products.
d) Partial hydrolysis of the peptide followed by chromatographic separation and sequence analysis yielded the following di- and tripeptides.
(Leu-Phe, Phe-Pro, Lys-Leu, Val-Lys, Val-Lys-Leu, Phe-Pro-Val, Pro-Val-Lys)
5.3.4 A2
Solution:
Amino acid composition: Equimolar amounts of Leu, Lys, Phe, Pro, and Val. This implies that each of these amino acids appears at least once in the peptide sequence.
Molecular weight: The molecular weight is approximately 1100 daltons. Assuming an average residue weight of 110 daltons, the peptide is likely composed of about 10 amino acids (1100/110 = 10).
Carboxypeptidase resistance: The peptide did not undergo hydrolysis by carboxypeptidase, which suggests:
The C-terminal residue is likely Pro (since Pro resists carboxypeptidase hydrolysis). Alternatively, the C-terminal residue may be blocked or modified, but since Pro is specifically resistant, it seems a more likely candidate. Edman degradation failure: The fact that no PTH (phenylthiohydantoin) products were yielded by Edman degradation suggests that the N-terminus is blocked, possibly by a cyclic structure or some other modification that prevents Edman sequencing.
Carboxypeptidase observation:
Since the peptide is resistant to carboxypeptidase, and Pro is known to resist this enzyme, the C-terminal residue is most likely Pro. Based on the sequence above, Pro is already positioned after Phe and is followed by Val-Lys, so this seems consistent with the enzyme resistance.
Edman degradation observation:
The N-terminal blockage could suggest that the N-terminus is involved in a modification or a cyclic structure. However, based on the fragments we have, Val appears to be the first residue, but it might be modified in some way.
piece the fragments together like a puzzle of overlapping sequences
Val-Lys-Leu Phe-Pro Pro-Val-Lys
\ / \ / \
Leu-Phe Phe-Pro-Val Lys-Leu
Unused: Val-Lys Most likely sequence is Val-Lys-Leu-Phe-Pro-Val-Lys
From b), since the entire peptide is 1100 dalton and each amino acid has 110 daltons, we can guess there are only 10 amino acids.
Estimate the pI of the peptide in the problem above.
Solution:
| pI | N-term | Lys | Lys | C-term | Net |
|---|---|---|---|---|---|
| 9 | 0 | +1 | +1 | -1 | +1 |
| 7 | +1 | +1 | +1 | -1 | +2 |
| 10 | 0 | +0.5 | +0.5 | -1 | 0 |
pI is 10
6 Research
6.1 Selecting stocks
vFemale x Male
\(\frac{+}{+} ; \frac{\alpha\text{1-Split-GFP/+}}{+} \times \frac{IF}{CyO} ; \frac{TM3}{TM6}\)
Select for GFP+Male CROSS Female
\(\frac{+}{\text{CyO or IF}} ; \frac{\alpha\text{1-Split-GFP/+}}{\text{TM6 or TM3}} \times \frac{IF}{\underset{w+}{CyO-Cre}} ; \frac{TM3}{TM6}\)
Select for White-eyed mutant aka \(w^+\) which is caused by \(\text{CyO-Cre}\)
CounterSelect \(\frac{TM3}{TM6}\) and \(IF\)\(\frac{+}{\text{Cyo-Cre}} ; \frac{\alpha\text{1-Split}}{\text{TM6 or TM3}} \times \frac{+}{\text{Cyo-Cre}};\frac{\beta\text{1-Split}}{\text{TM6 or TM3}}\)
CounterSelect \(\frac{TM3}{TM6}\) and \(w^{+} (\text{Cyo-Cre})\)vFemale CROSS Male
\(\frac{+}{+} ; \frac{\alpha\text{1-Split}}{\beta\text{1-Split}} \times \frac{\text{UAS-CD8-GFP}}{+}\)
Select for \(w^+\)
Select for \(GFP^+\)
- Male CROSS Female
\(\frac{+}{+} ; \frac{\alpha 1 \times \beta 1}{\text{UAS-CD8-GFP}} \times \frac{TM3}{TM6}\)
CounterSelect \(w^+\)
- \(\frac{+}{+} ; \frac{\alpha 1 \times \beta 1}{\text{TM3 or TM6}}\)
6.2 Protein Quantification
- Separate heads
- Homogenize in RIPA + P.I. (100: 1)
- 20 min on Ice incubatation
- Lysis
- Grind in tubes for 15 minutes
- Add SP
- Gel then Transfer
